[BI] Key Terms in T2T Genome Assembly and Pangenomics
This post summarizes essential terms and techniques used in Telomere-to-Telomere (T2T) genome assembly and pangenome research, with concise definitions and reference illustrations
What are the T2T assembly and Pangenome?
T2T (Telomere-to-Telomere) assembly refers to a complete, gapless genome assembly from one telomere to the other, including previously unresolved regions like centromeres and rDNA.
Pangenome is a comprehensive reference model that captures the full range of genomic diversity across individuals, populations, or species—going beyond a single linear reference.
🖥️ Sequencing & Assembly Techniques in T2T & Pangenomics
| Technique | Definition | Purpose / Strengths |
|---|---|---|
| HiFi (PacBio High-Fidelity) | Long-read sequencing using circular consensus sequencing (CCS) to achieve high per-base accuracy (~Q30) with read lengths of ~15–25 kb. | Long reads with high accuracy; ideal for resolving repeats and phasing haplotypes. |
| ONT (Oxford Nanopore) | Long-read sequencing using nanopores that measure changes in electrical current as DNA passes through. | Ultra-long reads (100 kb~Mb); great for spanning entire repeats, centromeres, telomeres. |
| Bionano Optical Mapping | Maps ultra-long DNA molecules (100 kb–Mb) by fluorescently labeling specific motifs and imaging them in nanochannels. | Scaffolding assemblies, detecting large structural variants (SVs), validating contiguity. |
| Hi-C (Chromatin conformation capture) | Captures 3D spatial proximity between DNA regions using proximity ligation and sequencing. | Helps in chromosome-scale scaffolding; anchors contigs based on nuclear organization. |
| Strand-seq | Single-cell sequencing technique that sequences only one strand of DNA (template strand). | Detects inversions, sister chromatid exchange; critical for resolving orientation and phasing in T2T. |
| Phasing | Determining which variants are inherited together on the same chromosome. | Enables haplotype-resolved genome assemblies; critical for understanding allele-specific variation. |
| Trio-binning | Method for separating reads by parental origin using sequencing data from both parents. | Produces haplotype-resolved assemblies from diploid individuals. |
| k-mer | Substrings of length k from a nucleotide sequence (used in assembly, mapping). | Fundamental unit for genome assembly, error correction, repeat detection, and sequence comparison. |
Comparison of Genomic Technologies
| Technology | Read Length | Accuracy | Ideal for |
|---|---|---|---|
| HiFi | 15–25 kb | High (~Q30+) | Resolving repeats, phasing |
| ONT | 100 kb–1 Mb | Moderate | Centromeres, telomeres |
| Bionano Optical Mapping | 100 kb–Mb (molecule length) | N/A | SV detection, scaffolding |
| Hi-C | N/A (proximity-based) | N/A | Chromosome scaffolding |
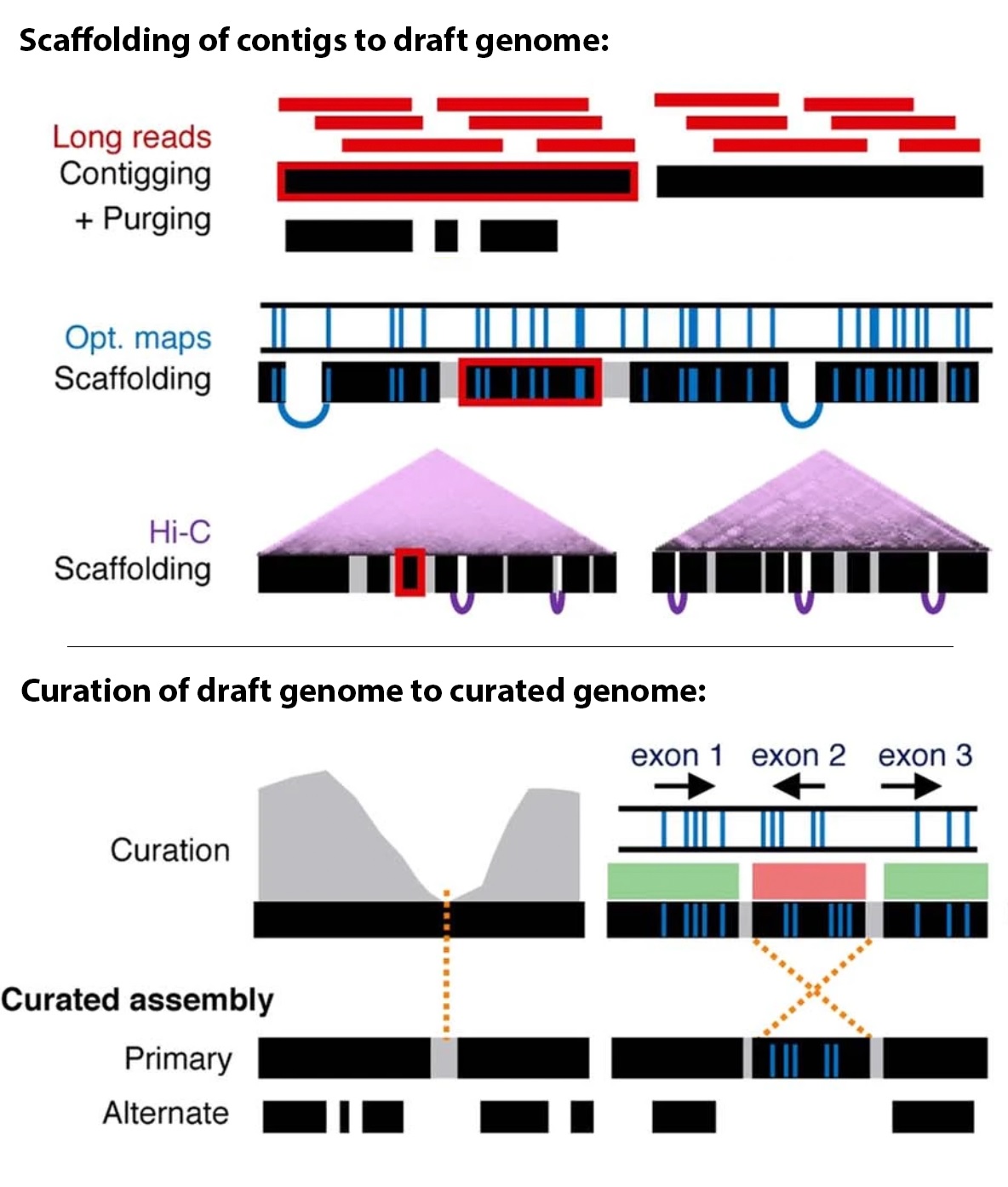
🧬 Key Genomic Regions and Concepts in T2T and Pangenome Research
| Term | Description | Why It Was Difficult to Assemble |
|---|---|---|
| rDNA repeats | Repeated ribosomal RNA gene clusters; essential for protein synthesis. | Highly repetitive sequences made alignment and assembly ambiguous. |
| Acrocentric DNA | DNA on short arms of acrocentric chromosomes (13, 14, 15, 21, 22) containing rDNA and satellite DNA. | Dense with similar repeats; often collapsed in assemblies. |
| Segmental Duplications (SDs) | Large blocks (1–200 kb) of duplicated DNA with high sequence similarity. | Difficult to distinguish between duplicated regions. |
| Centromeres | Central chromosome regions critical for proper segregation during cell division; rich in alpha-satellite repeats. | Extremely repetitive and megabase-sized; unreadable with short reads. |
| Telomeres | Chromosome ends composed of repetitive TTAGGG sequences; protect against genomic instability. | Simple and extremely repetitive; hard to resolve start/end. |
| VNTRs (Variable Number Tandem Repeats) | Short sequence motifs repeated in tandem with variable counts among individuals. | Hard to accurately estimate repeat number using short reads. |
| Heterochromatin | Densely packed, gene-poor chromatin regions, often transcriptionally silent. | Largely inaccessible and repetitive; often excluded from reference genomes. |
| Satellite DNA | Tandemly repeated sequences found in centromeres and heterochromatic regions. | Similar to alpha-satellite; hard to sequence and map due to size and uniformity. |
| Polymorphic Structural Variants (SVs) | Large-scale genome changes (insertions, deletions, inversions, duplications) that vary among individuals. | Often missed or mischaracterized by short-read sequencing. |
| Inversion toggling | Regions that repeatedly flip orientation between species or populations. | Hard to detect with short reads; can be misassembled. |
| Haplotypes / Haplotype-resolved assembly | Distinct sets of variants inherited together on a chromosome. | Phasing across long distances needs long reads and complex algorithms. |
| Haplotype false duplication | Error where heterozygous variants are misrepresented as duplicated regions in assembly. | Caused by failure to collapse allelic variation; often occurs in diploid genomes assembled without phasing. |
| Microchromosomes | Tiny, gene-rich chromosomes seen in birds, reptiles, etc. | Numerous, small, GC-rich, and repeat-dense; often fragmented or misassembled due to their short length and limited long-range scaffolding signals. |
🐒 Structural & Evolutionary Genomics Concepts
| Term | Definition |
|---|---|
| Hybridization | Breeding between genetically distinct lineages. |
| Introgression | Gene flow between species/populations via hybrid backcrossing. |
| Synteny | Conservation of blocks of genes across species, not necessarily in the same order. |
| Collinearity | Preservation of gene order across species. |
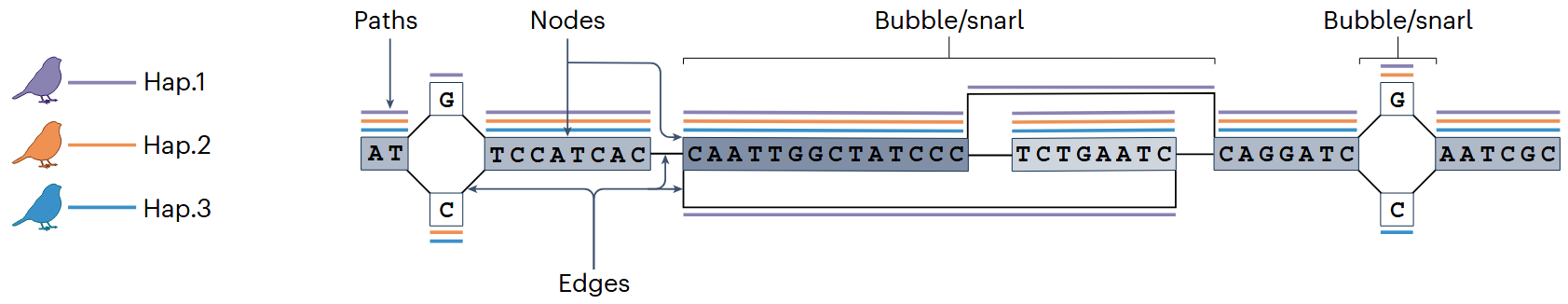
🧩 Pangenome Graph Concepts
| Term | Definition |
|---|---|
| Bidirected graph | Graph representing both DNA strands (forward/reverse). |
| Node | DNA sequence unit in the graph; may be shared across multiple haplotypes. |
| Edge | Connectors between nodes, indicating how they form complete sequences. |
| Path | A concatenation of nodes representing a specific haplotype. |
| Bubble | Region where paths diverge and reconverge, reflecting sequence variation. |
| Snarl | A generalized bubble with complex internal connections. |
Reference
-
Secomandi et al. (2024) Pangenome graphs and their applications in biodiversity genomics. Nature Genetics
-
Rhie et al. (2021) Towards complete and error-free genome assemblies of all vertebrate species. Nature
-
Rocha et al. (2024) Structural variation in humans and our primate kin in the era of telomere-to-telomere genomes and pangenomics. Curr Opin Genet Dev



Leave a comment