[BI] nf-core/fetchngs: Efficient NGS raw data download pipeline
In my previous post, I discussed several methods for downloading public NGS data, including the nf-core/fetchngs pipeline. This pipeline provides three retrieval options—FTP, Aspera, and sra-tools—and in today’s post, we’ll examine how each one differs in terms of performance and the types of errors you might encounter.
Retrieval Methods in nf-core/fetchngs
- FTP (default)
- Aspera (
--download_method aspera) - sra-tools (
--download_method sratools)
When using FTP or Aspera, compressed FASTQ files (fastq.gz) are fetched from the ENA database. If you opt for the sra-tools method, sra files are downloaded from the SRA database and then converted into FASTQ format.
Performance
I compared the three methods by measuring download speeds for four datasets—two single-end and two paired-end—each of similar size. Because the sra-tools approach involves first downloading .sra files and then converting them to .fastq, it ended up taking relatively more time. In scenarios with smaller datasets, FTP turned out to be somewhat quicker, but for large-scale downloads, the Aspera method consistently delivered faster performance.
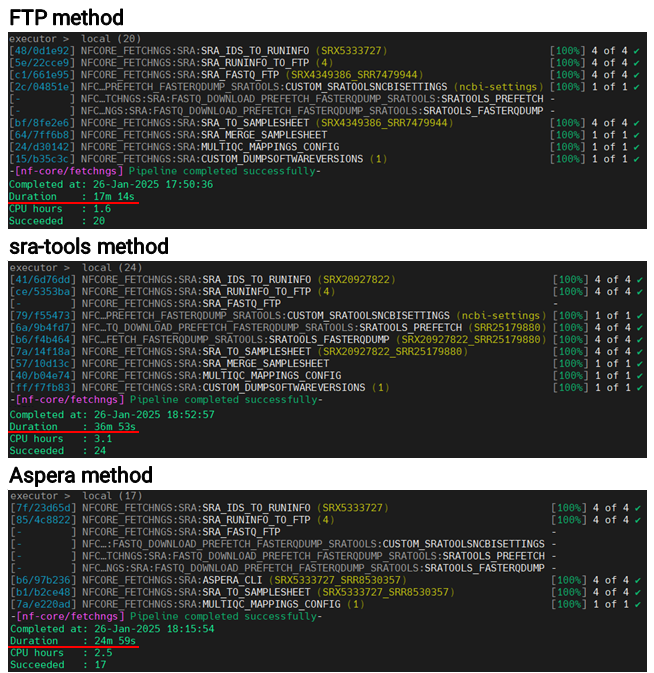
Errors
I tried all the methods, but I frequently ran into errors. The FTP and sra-tools methods produced time limit errors (20 minutes and 4 hours, respectively) due to the set time constraints. With fixed time limits like these, I concluded that downloading large files would be difficult. As for the Aspera method, I’m not entirely sure what caused the error, but I suspect unstable network connectivity might be to blame.
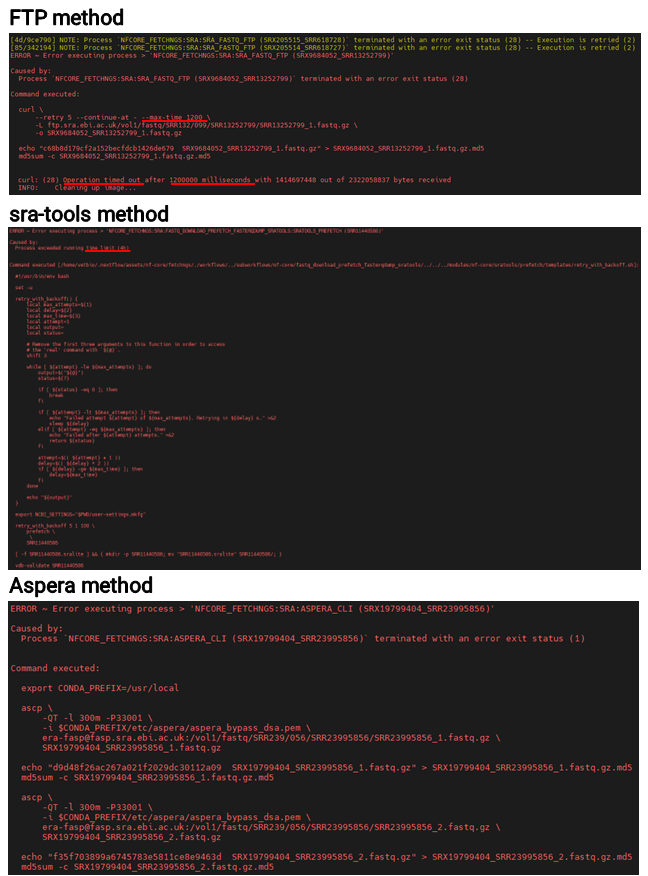
What’s the best method
I experienced the most errors with the Aspera method, but it also provided the fastest download speeds. By repeatedly running the pipeline with the -resume option, I was ultimately able to retrieve all the data. As shown in the example below, I ran the pipeline in a for loop to ensure every dataset was downloaded. For any items that still failed under Aspera, switching to FTP or sra-tools successfully completed the process.
for i in {1..50};do
echo "!! Iteration: "$i
nextflow run nf-core/fetchngs \
-profile singularity \
--input ids.csv \
--outdir 00_raw \
-r 1.12.0 \
--download_method aspera \
-resume
done
Additional Information
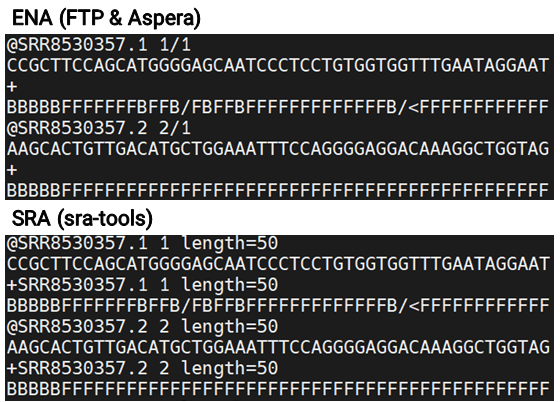
While comparing the methods, I noticed that file sizes differed between the ENA and SRA downloads. Curious whether this meant there were differences in the actual content, I checked the line counts, which turned out to be the same.
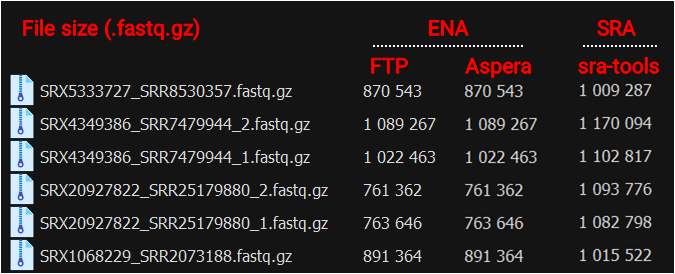
Upon further review, I confirmed that all key information matched, so there were no issues for analysis. Interestingly, the files from ENA were about 20% smaller on average, suggesting they could be more efficient for storage management.




Leave a comment