[Exploration] Primate 2: Genome Characteristics (Size, Karyotype, Assemblies)
Primates are a diverse mammalian order includes humans and 500+ living species—from ~30 g mouse lemurs to >200 kg gorillas. Building on Part 1 (phylogeny and distribution), this post shifts to the molecular view: genome size, karyotype evolution, and reference assemblies.
Genome size
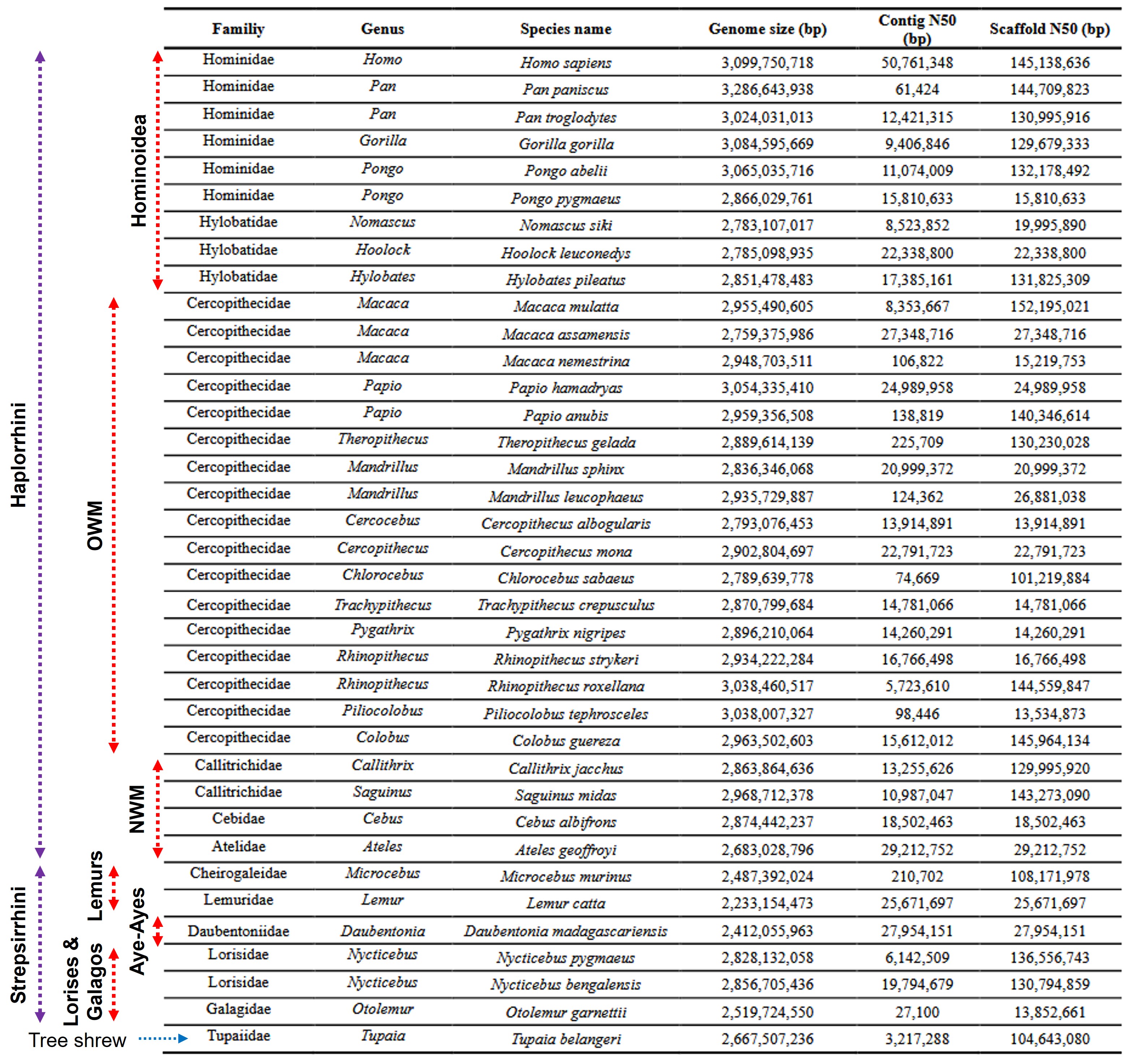
Primate genomes generally are on the larger side among mammals. Across the order, assembled sizes cluster around ~3.0 gigabases (Gb), very close to human [1].
- Apes (Hominoidea):
- Human (Homo sapiens): ~3.10 Gb.
- Great apes (Pan, Gorilla, Pongo): 2.87–3.29 Gb (avg ~3.07 Gb).
- Gibbons (Hylobatidae): 2.78–2.85 Gb (avg ~2.81 Gb; smallest among apes).
- Old World monkeys (Cercopithecoidea): 2.76–3.05 Gb (avg ~2.92 Gb).
- New World monkeys (Platyrrhini): 2.68–2.97 Gb (avg ~2.85 Gb).
- Strepsirrhines:
- Lemurs (Lemuriformes/Chiromyiformes): 2.23–2.49 Gb (avg ~2.38 Gb; clearly smaller).
- Lorises & galagos (Lorisiformes): 2.52–2.86 Gb (avg ~2.73 Gb; broadly comparable to monkeys).
- Outgroup context: Tree shrew (Scandentia) ~2.67 Gb.
Reading the table: “Genome size (bp)” reports assembled length. Contig N50 and Scaffold N50 summarize contiguity—higher values usually indicate long-read and/or Hi-C–assisted builds.
PRIMATE EVOLUTION - MAJOR GROUPS ONLY
├─ SCANDENTIA (Tree Shrews)
│ └─ 1 species • 2.67 Gb
│
└─ PRIMATES
│
├─ STREPSIRRHINI (Prosimians)
│ ├─ LEMURIFORMES (Madagascar Lemurs)
│ │ ├─ 3 species • 2.23-2.49 Gb
│ │ └─ Average: 2.38 Gb
│ └─ LORISIFORMES (Lorises & Galagos)
│ ├─ 3 species • 2.52-2.86 Gb
│ └─ Average: 2.73 Gb ★ Comparable to other primates
│
└─ HAPLORHINI
├─ PLATYRRHINI (New World Monkeys; NWM)
│ ├─ 4 species • 2.68-2.97 Gb
│ └─ Average: 2.85 Gb
│
└─ CATARRHINI
├─ CERCOPITHECOIDEA (Old World Monkeys; OWM)
│ ├─ 17 species • 2.76-3.05 Gb
│ └─ Average: 2.92 Gb
│
└─ HOMINOIDEA (Apes)
├─ HYLOBATIDAE (Lesser Apes)
│ ├─ 3 species • 2.78-2.85 Gb
│ └─ Average: 2.81 Gb ★ Smallest
│
└─ HOMINIDAE (Great Apes)
├─ OTHER GREAT APES
│ ├─ 5 species (Pan, Gorilla, Pongo)
│ ├─ Range: 2.87-3.29 Gb
│ └─ Average: 3.07 Gb
│
└─ HOMO SAPIENS ─ 3.10 Gb
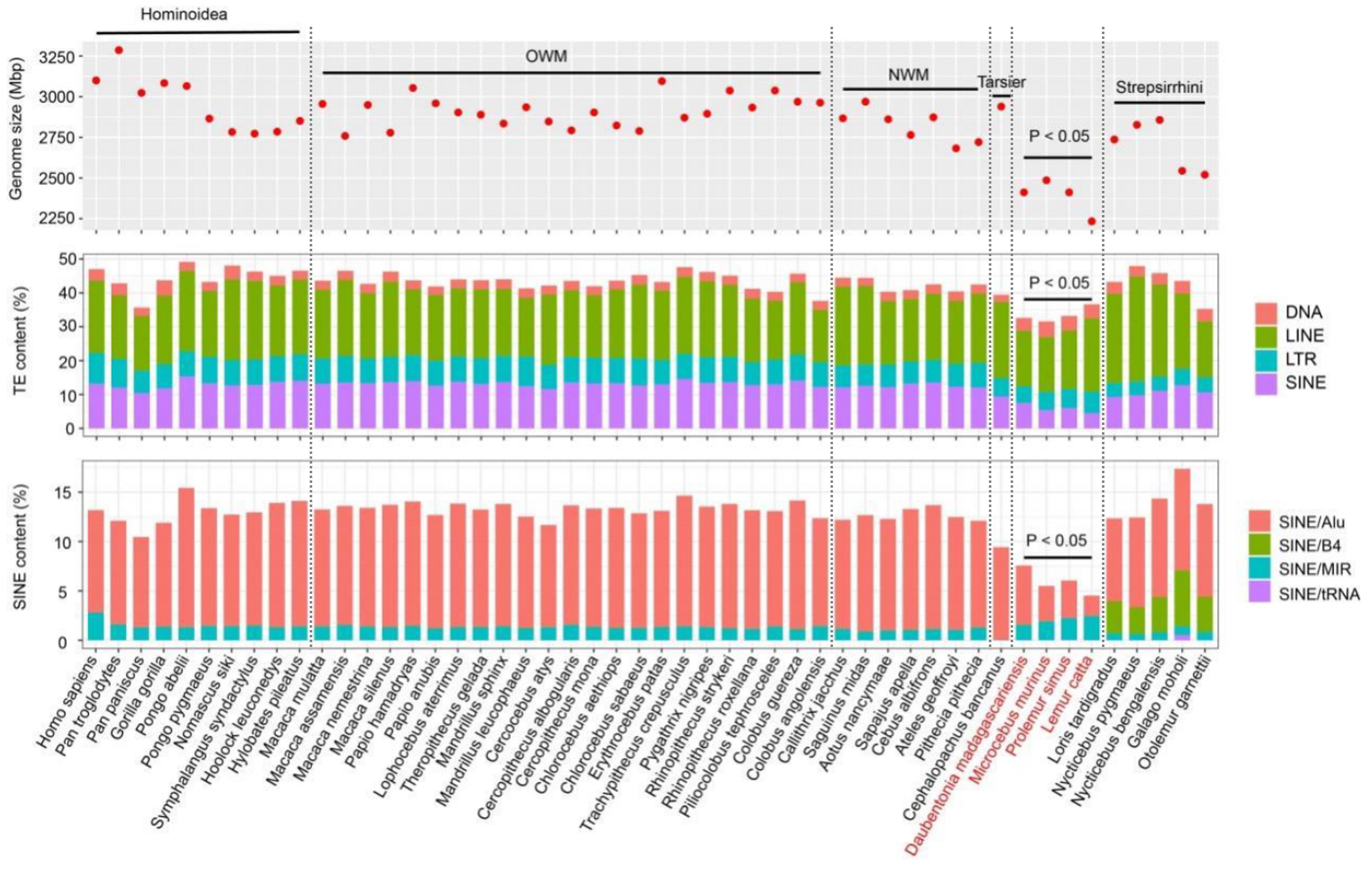
Why are simian genomes larger?
Across primates, differences in genome size largely track transposable elements (TEs) rather than gene counts. In Simiiformes, assembly length increases are driven by SINE expansions—especially Alu (~11% of the human genome), which inflate introns and intergenic regions. Lemurs carry far fewer SINEs (Alu ~3.9%), matching their smaller assemblies. Alu activity shows lineage-specific bursts: ~40–45 Ma in Simiiformes (dominated by AluS/AluSx) and ~34–39 Ma in Lorisiformes (AluJ/AluJb).
Mechanism & consequences. Alu retrotransposition copies and pastes short sequences across the genome, increasing bulk DNA and creating substrates for non-allelic homologous recombination (structural variation). Some insertions are later exapted as regulatory elements, but the primary effect on size is simple sequence accretion.
Bottom line: Simian genomes trend larger because they experienced historical waves of Alu expansion, while lemur lineages—lacking comparable Alu loads—retain smaller assemblies despite similar numbers of protein-coding genes.
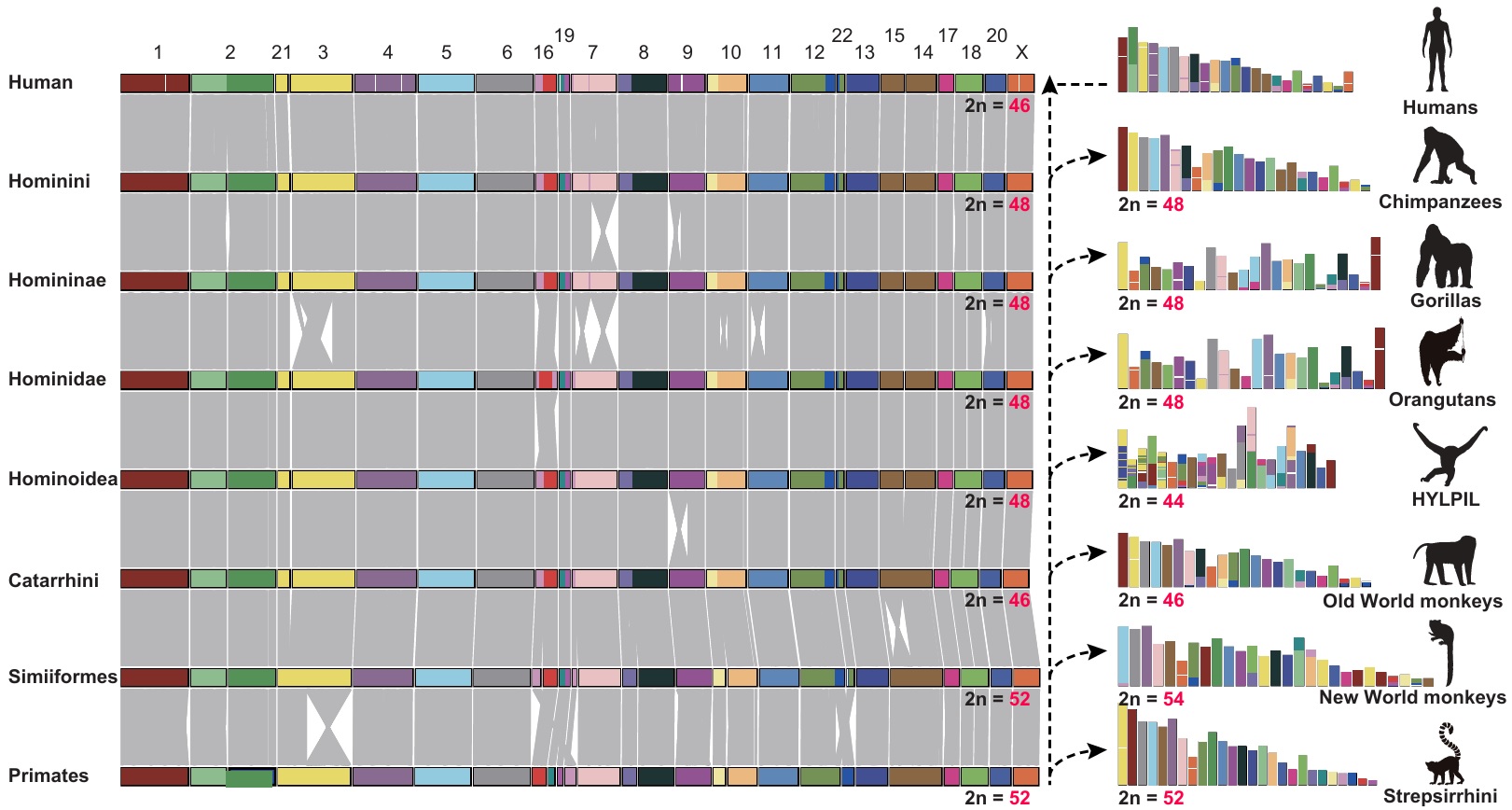
Chromosome and karyotype evolution
Primate chromosomes are highly syntenic—large blocks of gene order are conserved—yet their counts have shifted through time mainly via fusions and fissions (with inversions and translocations layered on top). Current ancestral reconstructions place the diploid number at 2n = 52 for both Primates and Simiiformes (revising earlier 50), consistent with an early chromosome-8 fission. Downstream, Catarrhini is inferred at 2n = 46, and Hominoidea at 2n = 48, steps that explain much of today’s variation.
In living taxa, humans carry 2n = 46 owing to the well-known fusion that formed chromosome 2, whereas other great apes (chimpanzee, gorilla, orangutan) retain 2n = 48. Gibbons show additional lineage-specific changes (2n = 44). Old World monkeys typically fall near 42–46, while New World monkeys often have the highest numbers (frequently 2n = 54). Strepsirrhines cluster around 2n = 52. Tallying large structural events along the path to humans shows an increasing rearrangement rate—about 2.38 events/Ma in Homininae and 5.56 events/Ma in Hominini.
Reference genomes
Primate diversity tops 500+ species, but high-quality reference genomes still cover only a fraction of that breadth. The snapshot below makes it clear [3-4]:
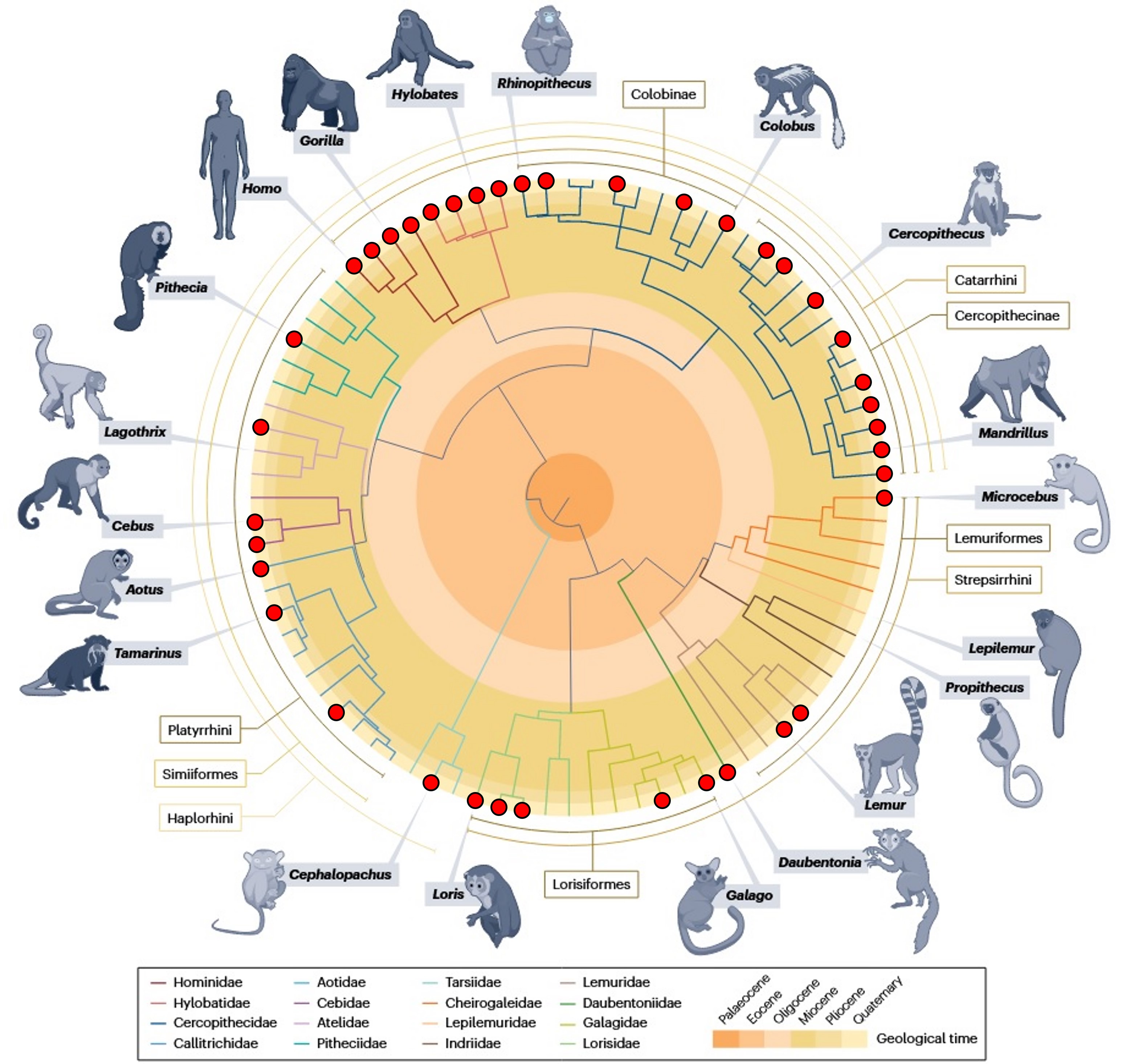
- Coverage today. About ~238 species have at least one assembly on public databases. Most of these are short-read, fragmented builds (left cluster in A) [2]. A smaller subset now has long-read assemblies with chromosome-scale contiguity, and only a handful reach near–telomere-to-telomere (T2T) quality; human T2T-CHM13 is the flagship example. B maps these assembly qualities across the primate tree—note how the best genomes are concentrated in a few clades.
- Why T2T/near-T2T matters. Long-read assemblies (PacBio HiFi + ultra-long ONT) reveal evolution that short reads miss. A recent eight-species survey spanning ~50 My of primate evolution showed ~27% of the genome affected by structural variation, and catalogued >1,600 large, highly divergent regions—over a third showing recurrent structural changes that touch hundreds of genes. This is the level of resolution needed to connect structure to adaptation.
- Breadth vs depth. Parallel efforts have delivered high-coverage short-read data for 233 species [3], which is excellent for variants, diversity, and dated phylogenies—but not a substitute for chromosome-level references when the goal is repeats, segmental duplications, TEs, and structural variants. The field now needs both: breadth for comparative context and T2T-grade references for each major lineage (especially under-sampled strepsirrhines and several New World monkey families).
Bottom line: reference coverage across primates is growing, but truly complete (near-T2T) assemblies are still rare. The next wave—T2T-quality genomes placed phylogenetically across the tree—will turn today’s broad catalog into a platform for high-resolution primate evolution.
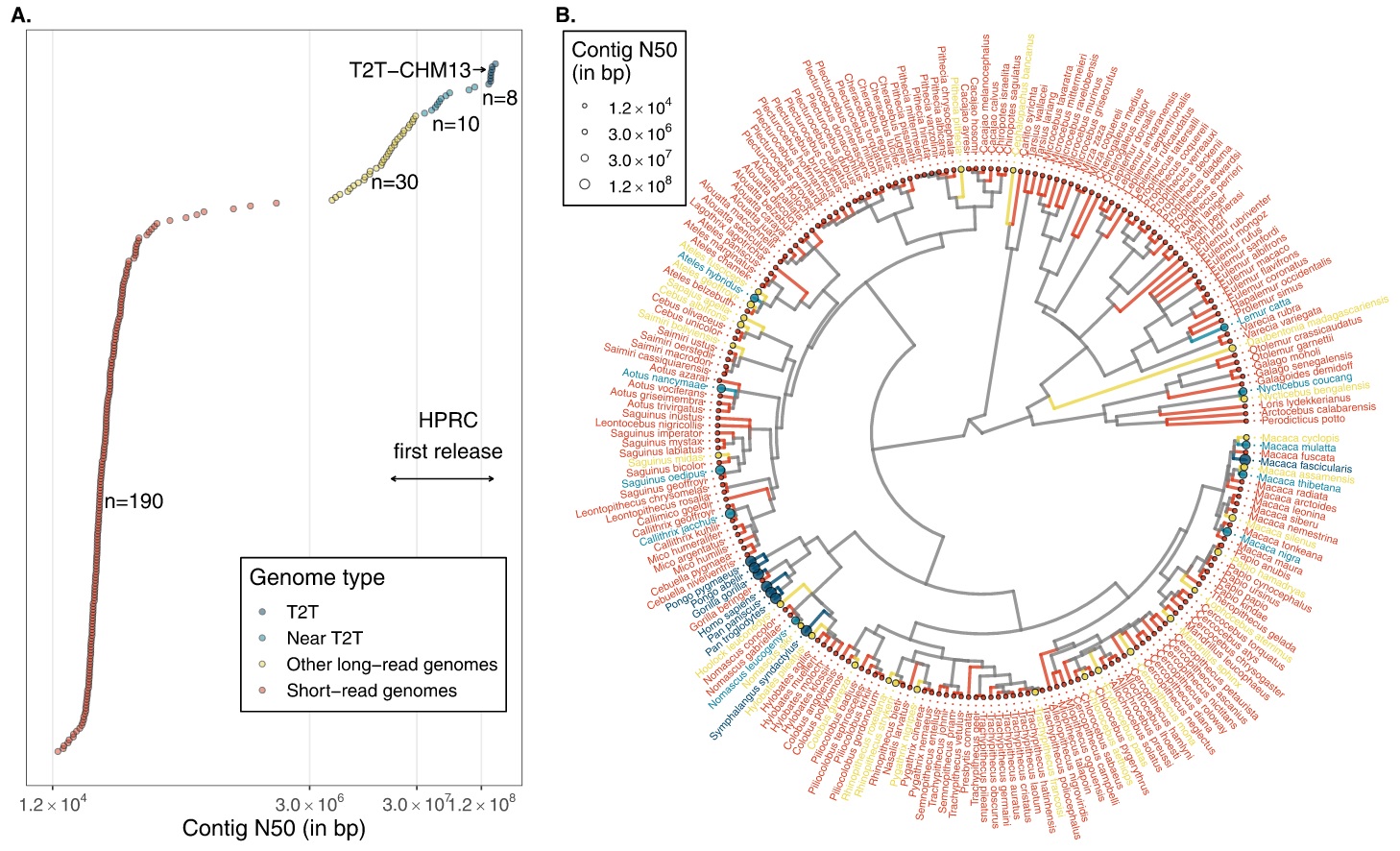
This time-calibrated tree of the 84 living primate genera flags branch tips with a ● where a high-quality reference genome exists. At a glance you can see coverage is strongest in apes (e.g., Homo, Gorilla, Hylobates), several Cercopithecidae genera (e.g., Rhinopithecus, Colobus, Cercopithecus, Mandrillus), and key Lemuriformes/Lorisiformes (e.g., Microcebus, Lemur, Propithecus, Lepilemur, Daubentonia, Galago, Loris). Missing circles mark gaps—under-sampled clades that are priorities for future near-T2T assemblies.
Reference
- Shao Y et al. Science. 2023.
- Rocha JL et al. Current Opinion in Genetics & Development. 2024.
- Kuderna LFK et al. Science. 2023.
- Roos C et al. Nature Reviews Biodiversity. 2025.



Leave a comment