[Exploration] Primate 1: Phylogeny and Geographic Distribution
Primates are a morphologically and ecologically diverse order of mammals distributed mainly across the tropical and subtropical regions. Depending on the taxonomy used, ~376–524 extant species are recognized, ranging in body sizes from the ~30 g Madame Berthe’s mouse lemur to >200kg eastern gorillas [1].
Primates in the tree of life
Primates are vertebrate mammals within the eukaryotic domain [2]: Eukaryotes → Animals → Vertebrates → Mammals → Primates
From the origin of life, primates sit on one slender branch of the eukaryotic tree: Life → Eukaryota → Opisthokonta → Animalia (Metazoa) → Bilateria → Deuterostomia → Chordata → Vertebrata (Craniata) → Gnathostomata → Osteichthyes (Euteleostomi/Teleostomi) → Sarcopterygii → Tetrapoda → Amniota → Synapsida → Mammalia → Theria → Eutheria (Placentalia) → Boreoeutheria → Euarchontoglires → Primates
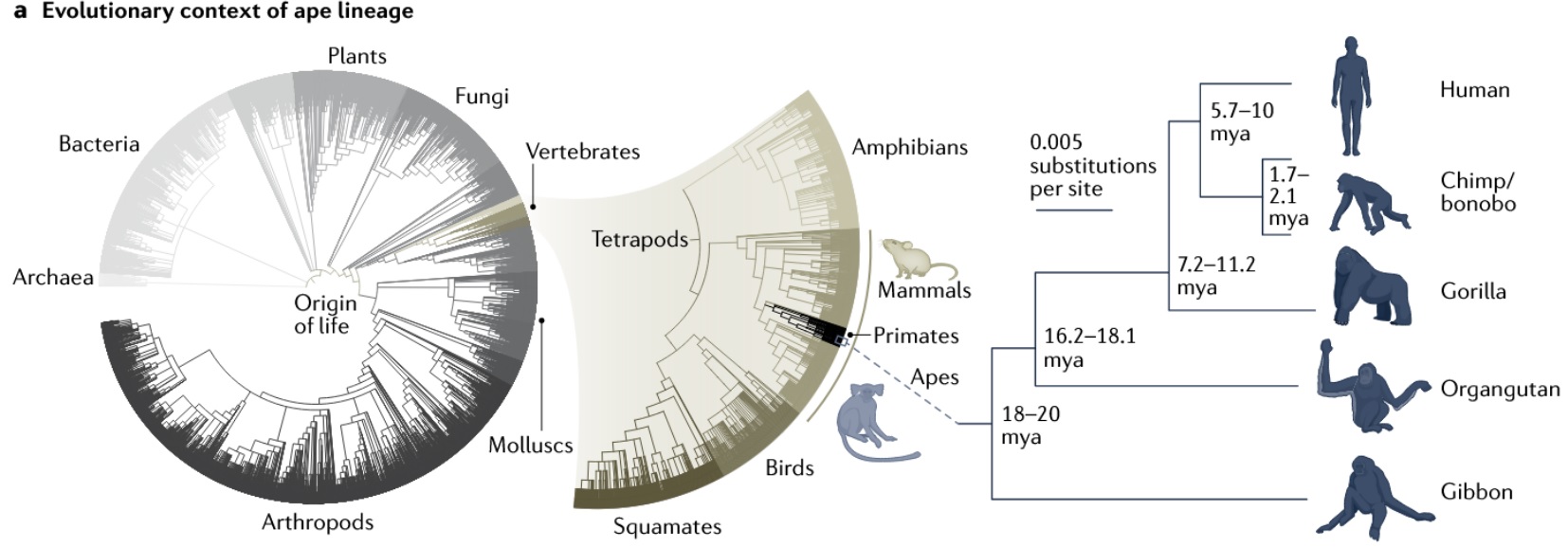
Major branches of primates
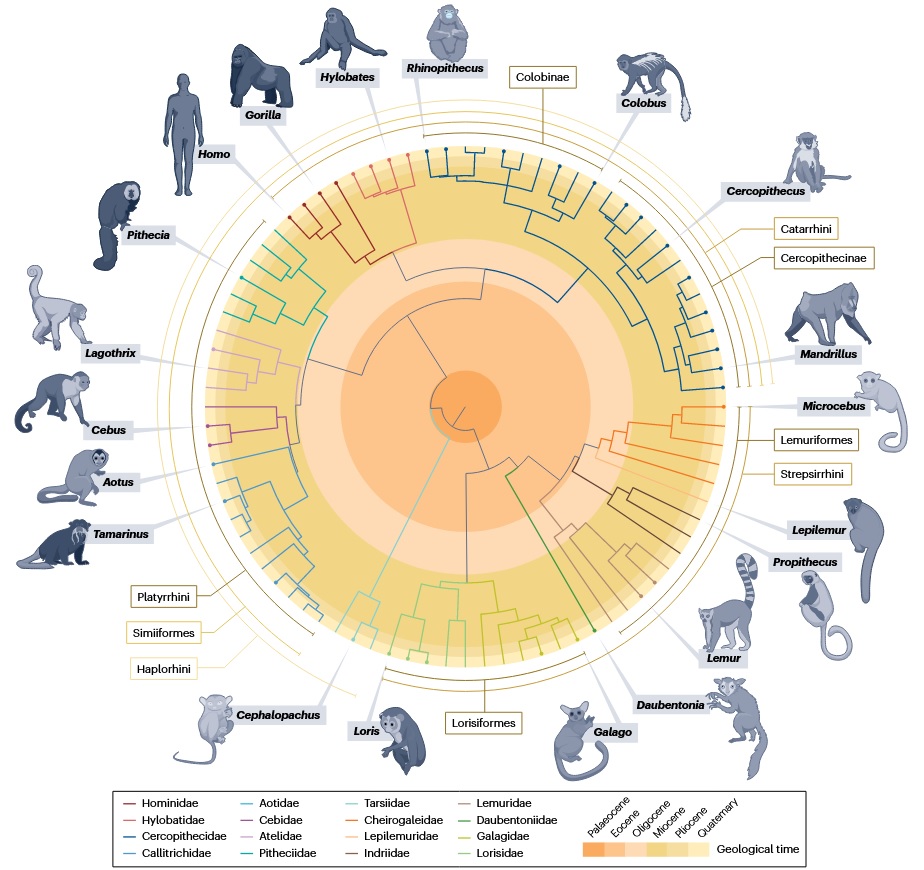
Living primates comprise two deep branches: Strepsirrhini (lemurs, lorises, galagos) and Haplorhini (tarsiers and simians). Simians further split into Platyrrhini (New World monkeys) and Catarrhini (Old World monkeys and apes) [1].
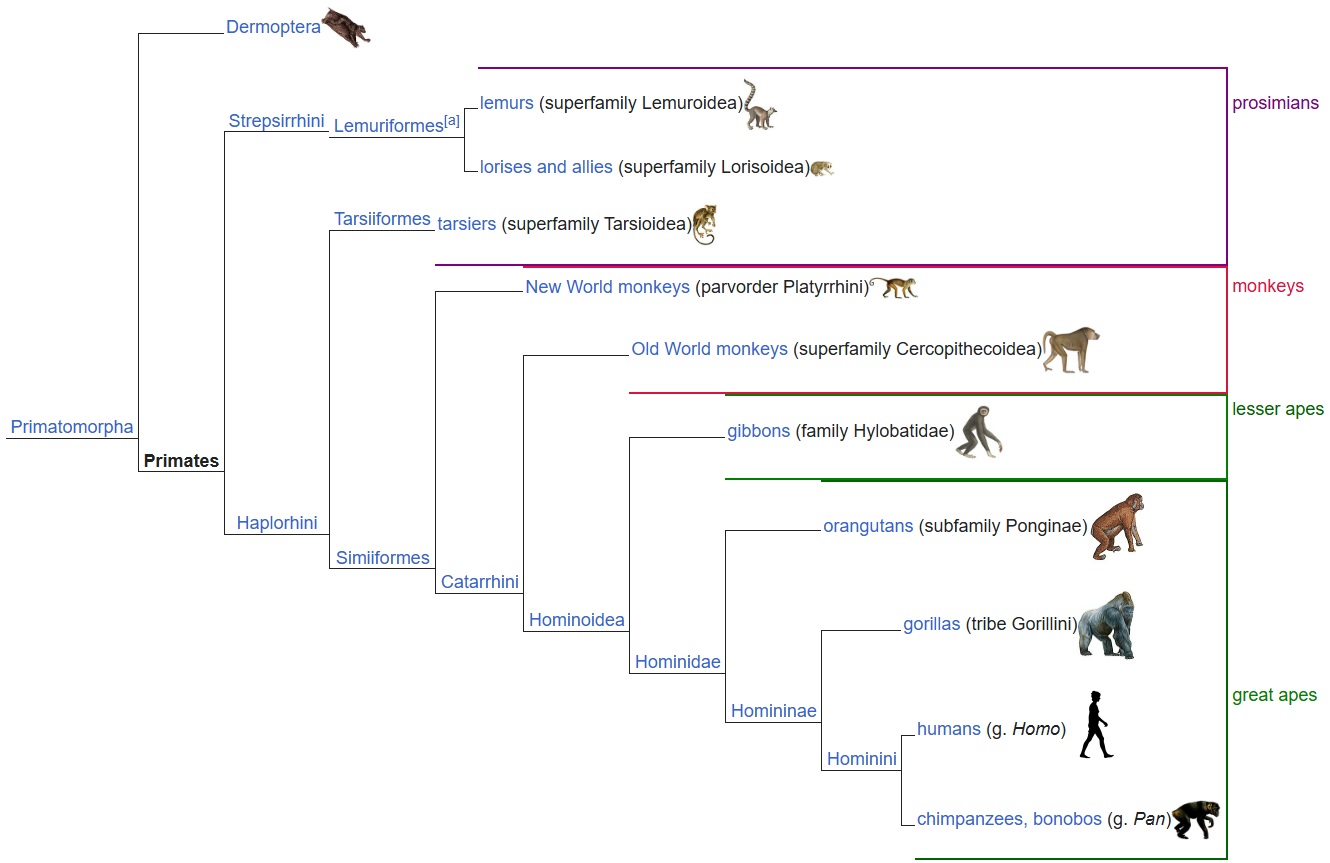
Building a robust primate phylogeny: two complementary approaches
1) De novo whole-genome references
- What it is: Generate new, high-contiguity reference assemblies (often long-read + Hi-C), then infer a species tree from whole-genome alignments of orthologous, syntenic sequence blocks.
- Strengths: Maximizes structural resolution (karyotypes, inversions/fusions), enables clean ortholog calls, detects lineage-specific duplications/TE bursts, and supports broad functional scans (selection, accelerated noncoding regions).
- Trade-offs: Costly and time-intensive; practical for fewer species, so breadth and within-species variation are limited relative to resequencing designs.
2) Cross-species resequencing
- What it is: Short-read whole-genome resequencing across many species (often multiple individuals/species), mapped to a curated set of references; infer a dated tree using conserved markers (e.g., UCEs).
- Strengths: Breadth and throughput—cost-effective coverage across the order; simultaneously yields genetic diversity, ROH, and demographic summaries; enables comparative estimates of mutation rates when combined with generation times.
- Trade-offs: Dependent on reference quality and cross-mapping choices; limited power for structural evolution compared with de novo assemblies.
Below is a summary of representative primate studies that applied these strategies.
Reference-genome–driven phylogenomics [3]
Study design & data
- Produced 27 new high-quality reference genomes and analyzed ~50 species total (new + published), emphasizing previously undersampled New World monkeys and strepsirrhines.
How the phylogeny is built
- Extracted ~433.5 Mbp of gap-free syntenic orthologous sequence, built whole-genome ML trees (ExaML, GTR+GAMMA), and dated nodes with fossil calibrations (MCMCtree).
Primary analyses & questions
- Karyotype reconstruction across primates; rates of rearrangement and hallmark events (e.g., fusions/fissions across key nodes).
- Genome-wide scans for positive selection, lineage-accelerated conserved elements, and gene-family expansions; highlights a surge of genomic innovation on the Simiiformes ancestor.
- Divergence times for crown primates and major subclades, with estimates near the K–Pg boundary for the primate MRCA.
Take-home: Depth per species enables structural and functional inferences that are hard to get from resequencing alone (karyotypes, synteny-aware selection tests).
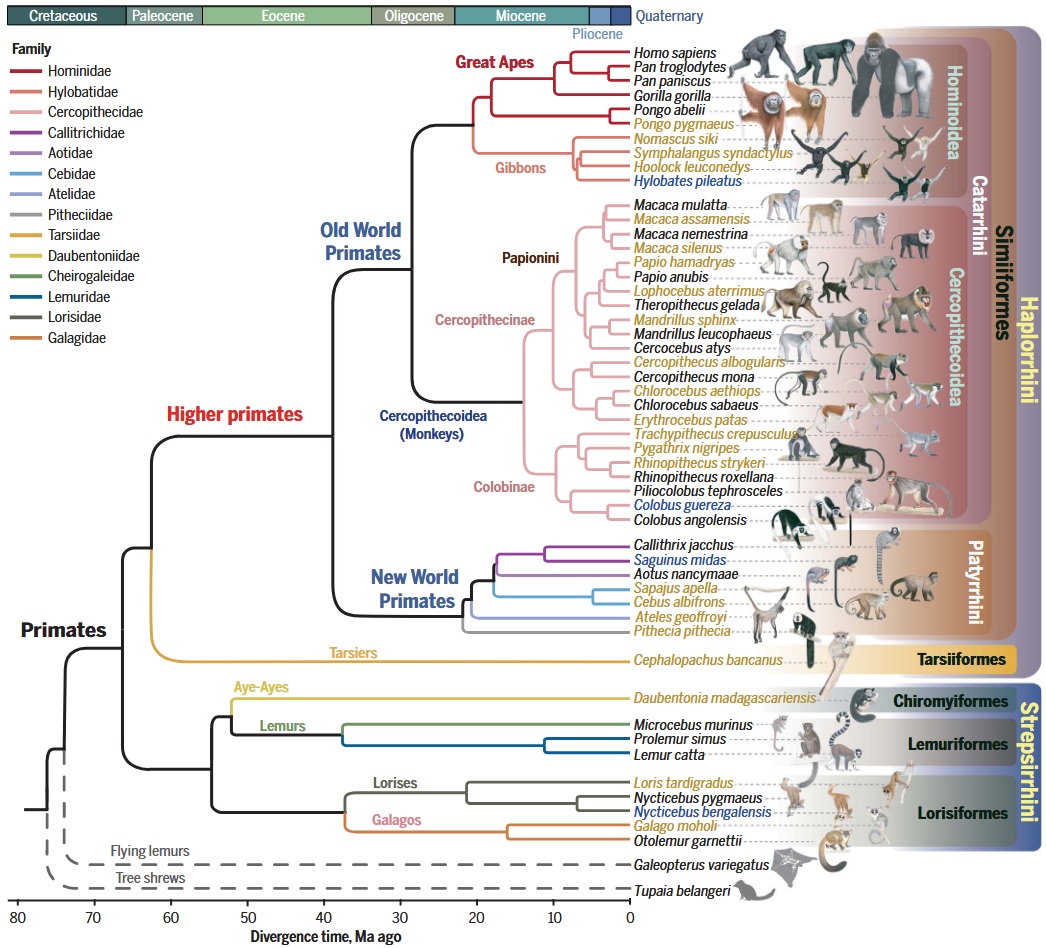
Cross-species resequencing atlas [4]
Study design & data
- High-coverage whole-genome data for 809 individuals from 233 species (covering 86% of genera and all 16 families), yielding a broad nuclear DNA phylogeny and updated divergence times.
How the phylogeny is built
- Identified ~3,500 ultraconserved elements (UCEs) + flanks across genomes; built per-locus ML trees and integrated them with a coalescent species tree, then fossil-calibrated the timeline.
Primary analyses & questions
- Genetic diversity (heterozygosity/ROH) across regions and families; diversity correlates with environment/sociality but not a simple predictor of extinction risk.
- Mutation rates: estimated μ per generation from substitution rates + generation time on the dated tree; μ correlates positively with generation time and is further shaped by effective population size (Ne) (drift-barrier signal).
- Human-relevance: cross-species catalogs to contextualize protein-changing variants and refine “potentially benign” vs pathogenic interpretations.
Take-home: Breadth and population-genetic readouts (heterozygosity, ROH, μ, Ne) at an unprecedented scale; ideal for questions about diversity, demography, and mutation-rate variation across the order.
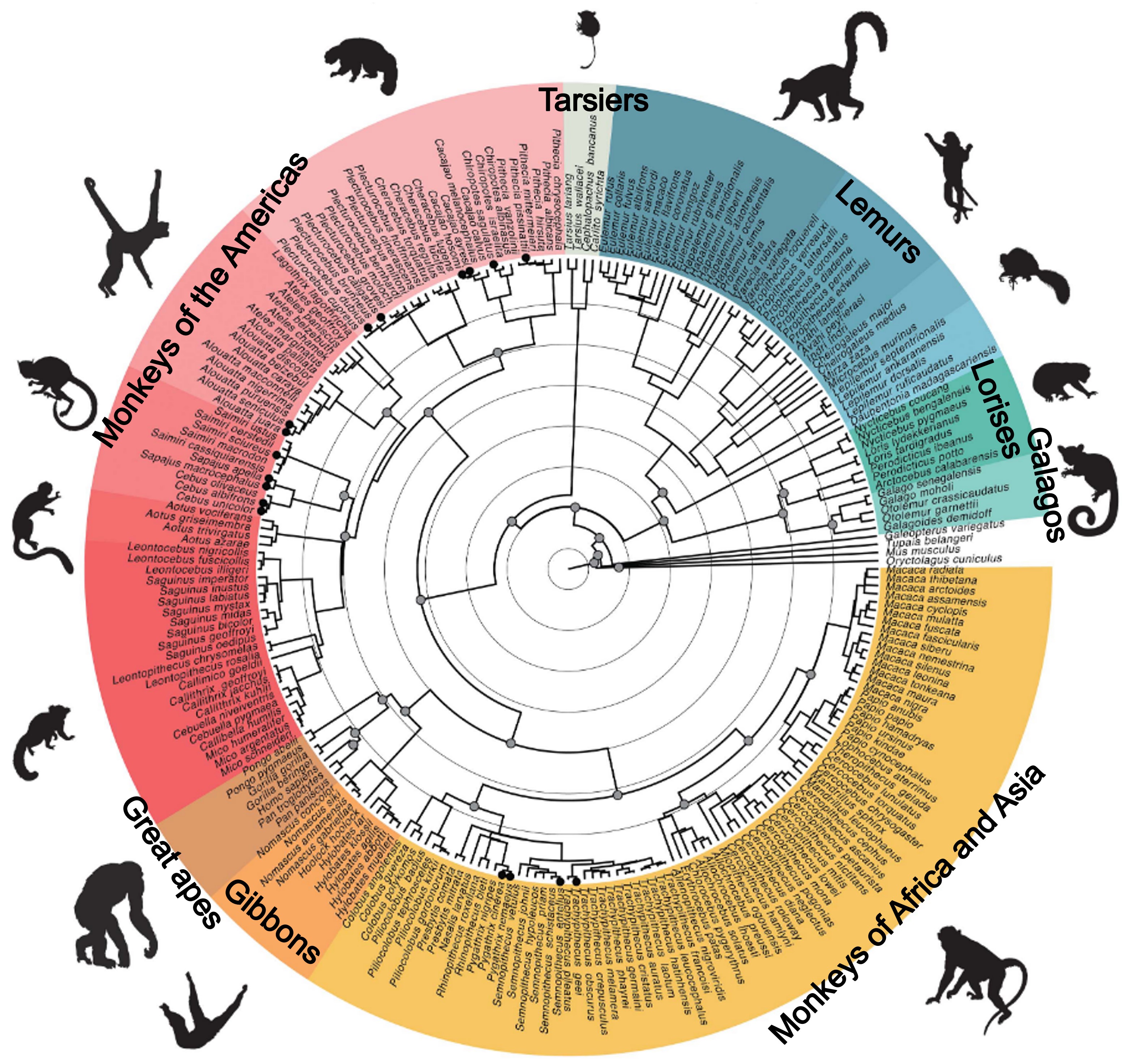
Figure below [5] shows a fossil-calibrated, time-scaled phylogeny for 84 extant primate genera. The backbone topology and divergence times primarily follow Kuderna et al. [4]; selected nodes and dates were recalculated using Shao et al. [3] and additional lineage-specific studies—not a simple merge of the two. Filled dots denote genera with high-quality reference genomes.
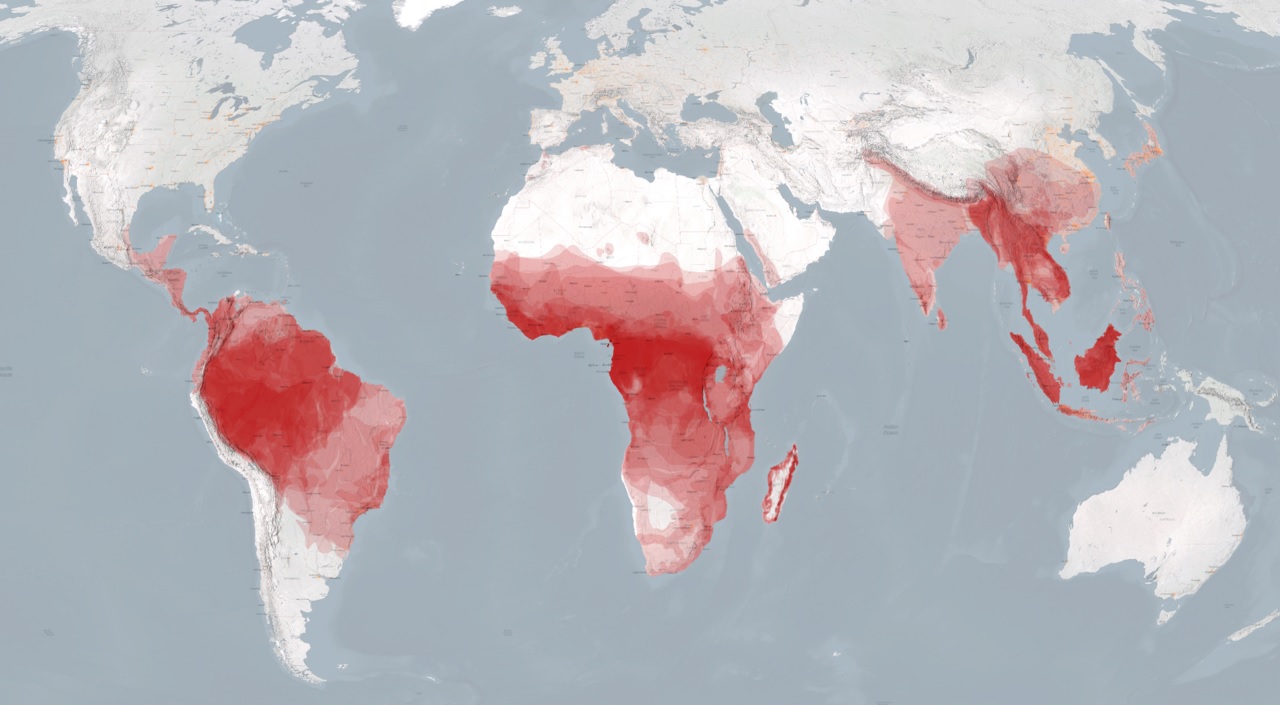
Geographical distribution (living primates)
Global range & density [1].
Living non-human primates concentrate in the tropics: the Neotropics (Central/South America), Afrotropics (sub-Saharan Africa, plus Madagascar), and Indo-Malaya (South/Southeast Asia). Hotspots are the Amazon Basin, Congo Basin, Madagascar, and Sundaland. They are largely absent from deserts and high latitudes, with only a few extra-tropical exceptions (e.g., some macaques).
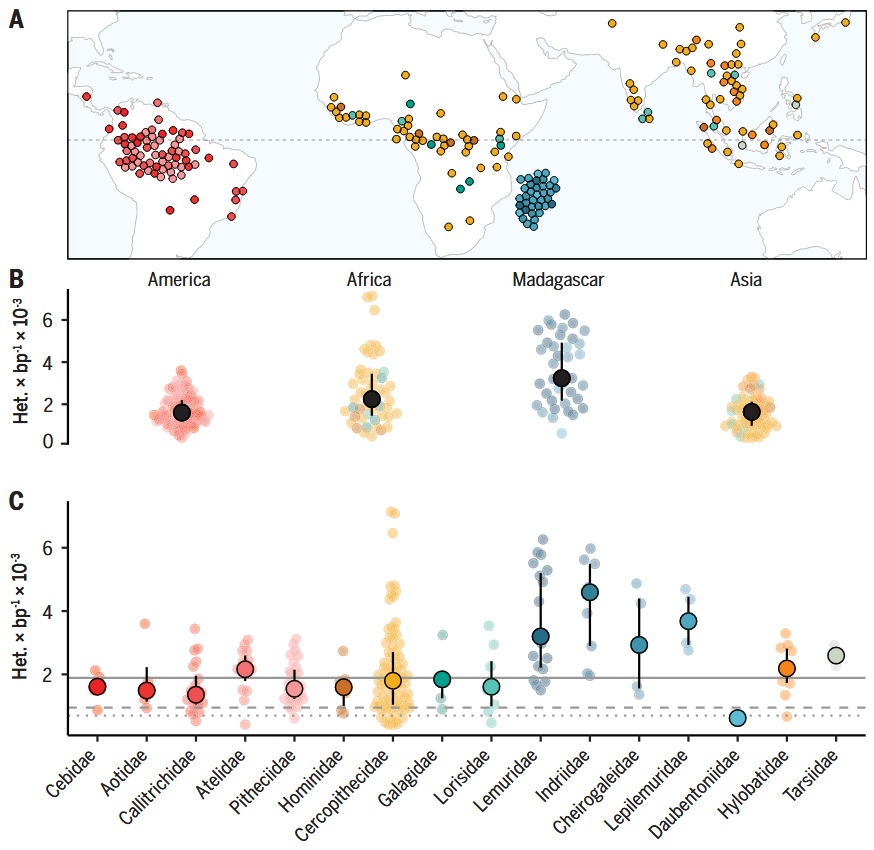
Continental and family patterns from a broad resequencing survey.
Kuderna et al. [4] sampled 809 individuals from 233 species. Sampling spans the Americas, Africa, Madagascar, and Asia (A). Within-species genetic diversity (heterozygosity) shows continent-level contrasts (B) and family-level spread (C):
- Neotropics (Cebidae, Aotidae, Callitrichidae, Atelidae, Pitheciidae): confined to the Americas; generally moderate diversity with between-family spread.
- Africa (Hominidae, Cercopithecidae, Galagidae, Lorisidae): wide geographic breadth; Cercopithecidae shows broad variance, Galagidae includes some high-diversity outliers.
- Madagascar (Lemuridae, Indriidae, Cheirogaleidae, Lepilemuridae, Daubentoniidae): island-endemic lineages with a wide range of diversity levels across families.
- Asia (Hylobatidae, Tarsiidae + Asian Cercopithecidae): concentrated in South/Southeast Asia; generally lower-to-moderate diversity, with family-specific exceptions.
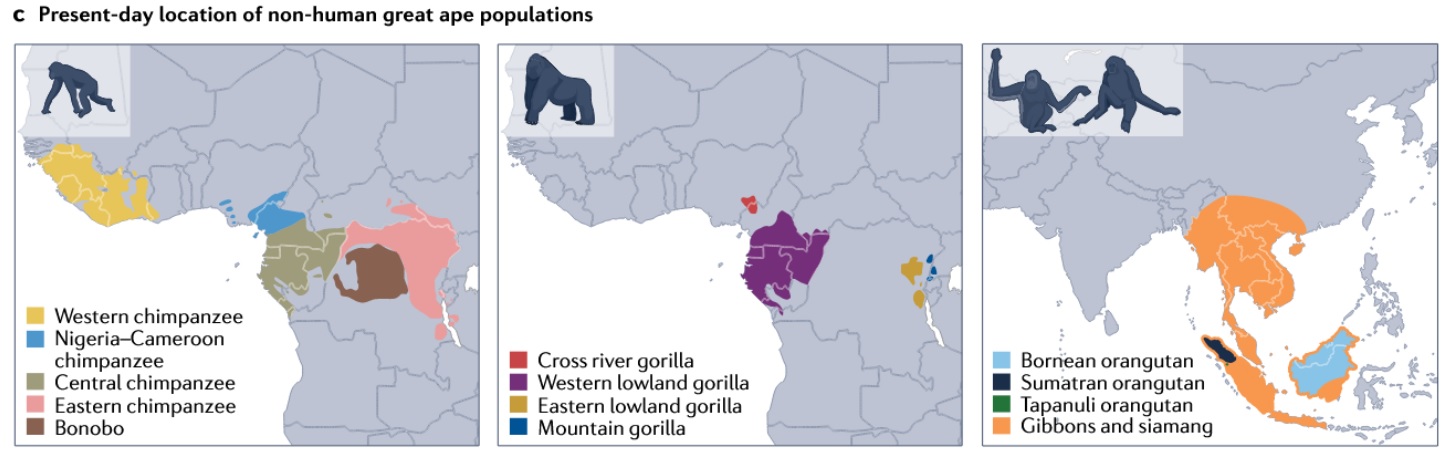
Lineage-specific present-day ranges [2].
Great apes provide a clear example of fine-scale distributions:
- Chimpanzees (four subspecies) and bonobos are confined to central and western Africa.
- Gorillas occur in west/central Africa (western lowland, Cross River) and eastern Africa (eastern lowland, mountain).
- Orangutans are island endemics in Southeast Asia (Borneo, Sumatra, Tapanuli).
- Gibbons and siamangs occupy mainland and insular Southeast Asia.
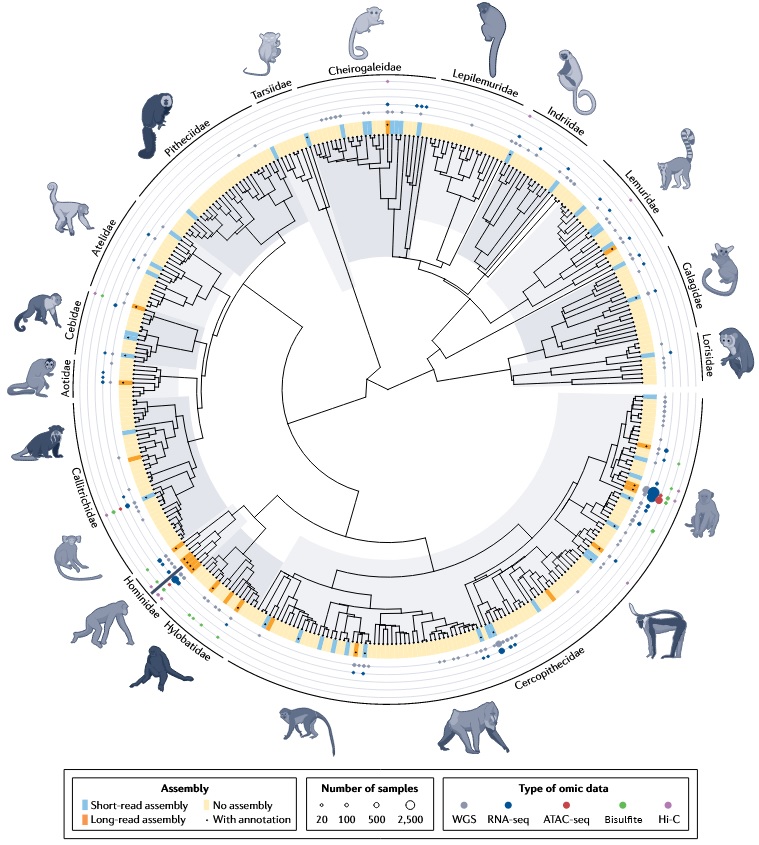
Primate omics resources on the phylogeny
Despite the large species richness, most high-quality resources—reference genomes, WGS, RNA-seq, ATAC-seq, Hi-C, methylomes—are clustered in a small set of model lineages (human and great apes; rhesus/cynomolgus macaques; marmoset) [6]. Coverage across many strepsirrhines and several New World monkey families remains sparse.
Why this matters:
- Dense data near humans → powerful for fine-scale evolutionary and disease inference in our closest relatives.
- Sparse data elsewhere → limits tests of trait evolution and comparative constraints across the full phylogeny; results can be human-centric or biased.
- Ongoing efforts are widening species coverage, but annotations and standardized phenotype metadata still lag behind.
Glossary
- Eukaryota: Cells with nuclei and membrane-bound organelles (animals, plants, fungi, protists).
- Opisthokonta: Major eukaryote clade that includes animals and fungi (and their unicellular relatives).
- Animalia (Metazoa): Multicellular, heterotrophic eukaryotes with specialized tissues—animals.
- Bilateria: Animals with bilateral symmetry and three germ layers (excludes sponges, cnidarians, ctenophores).
- Deuterostomia: Bilaterians in which the anus forms before the mouth in development; includes echinoderms and chordates.
- Chordata: Animals that, at some stage, have a notochord, dorsal hollow nerve cord, pharyngeal slits, and post-anal tail.
- Vertebrata (Craniata): Chordates with a skull and usually a vertebral column.
- Gnathostomata: Jawed vertebrates (all modern fishes and tetrapods).
- Osteichthyes (Euteleostomi/Teleostomi): “Bony vertebrates” — ray-finned fishes, lobe-finned fishes, and tetrapods.
- Sarcopterygii: Lobe-finned lineage within Osteichthyes; includes lungfish, coelacanths, and tetrapods.
- Tetrapoda: Four-limbed vertebrates (amphibians, reptiles, birds, mammals).
- Amniota: Tetrapods with an amniotic egg or equivalent membranes (reptiles, birds, mammals).
- Synapsida: The amniote lineage leading to mammals; defined by a single temporal opening in the skull.
- Mammalia: Hair, lactation, three middle-ear bones; includes monotremes, marsupials, and placentals.
- Theria: Mammals excluding monotremes; comprises Metatheria (marsupials) + Eutheria (placentals).
- Eutheria (Placentalia): Eutheria is the broader placental lineage; Placentalia refers to the crown group of living placental mammals.
- Boreoeutheria: A major placental clade uniting Euarchontoglires and Laurasiatheria.
- Euarchontoglires: Superclade combining Euarchonta (primates + colugos + treeshrews) and Glires (rodents + lagomorphs).
- Primates: The order comprising Strepsirrhini (lemurs, lorises, galagos) and Haplorhini (tarsiers + simians).
- Simians (Anthropoidea): Split into Platyrrhini (New World monkeys) and Catarrhini (Old World monkeys + apes).
- Apes (Hominoidea): Lesser apes (gibbons) and great apes (orangutans, gorillas, chimpanzees/bonobos, humans).
Reference
- Primate - Wikipedia
- Pollen AA et al. Nature Reviews Genetics. 2023.
- Shao Y et al. Science. 2023.
- Kuderna LFK et al. Science. 2023.
- Roos C et al. Nature Reviews Biodiversity. 2025.
- Juan D et al. Nature Reviews Genetics. 2023.



Leave a comment