[BI] Problematic regions and Mappability in genomic analysis
In my previous post, we delved into the concept of Blacklist regions in genomes. Expanding on thi...
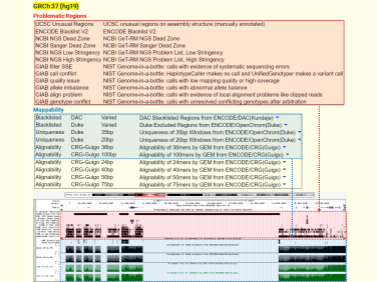
In my previous post, we delved into the concept of Blacklist regions in genomes. Expanding on thi...

Chromatin states in the field of epigenomics refer to the organization and modification of chroma...
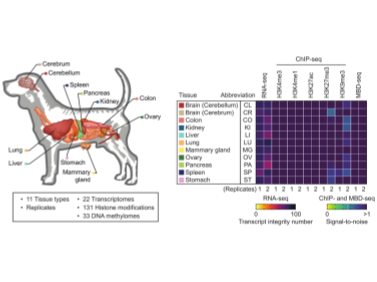
In this post, let’s explore what a blacklist is in the context of genomics and why certain region...
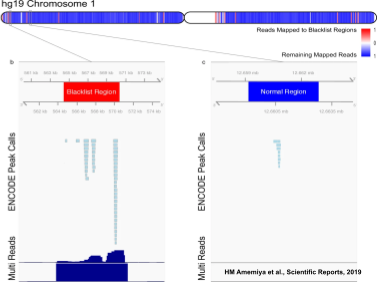
Types of Proteomic Data:
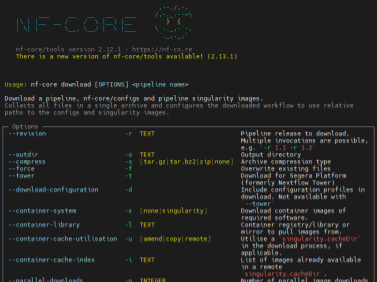
When running nf-core pipelines for the first time, I frequently encounter errors while pulling th...